Research
The primary research interest of the Laboratory of Computational Functional Genomics is the development of methods for the analysis and integration of omics data to understand underlying mechanisms of disease, facilitate generating testable hypotheses, and identify potential therapeutics in human and animal studies. Specifically, our research focuses on developing methods for inferring gene regulatory mechanisms, disease prediction, and discovering biomarkers based on genetic, genomic, epigenetic, and clinical data. Our approaches are diverse, including optimization, machine learning, and statistical modeling.
The following list highlights our recent research and collaborative work.
A. Recent computational tools for functional omics data
- ISEGnet Assessing comparative importance of DNA sequence and epigenetic modifications on gene expression using a deep convolutional neural network (Comput Struct Biotechnol J. 2022).
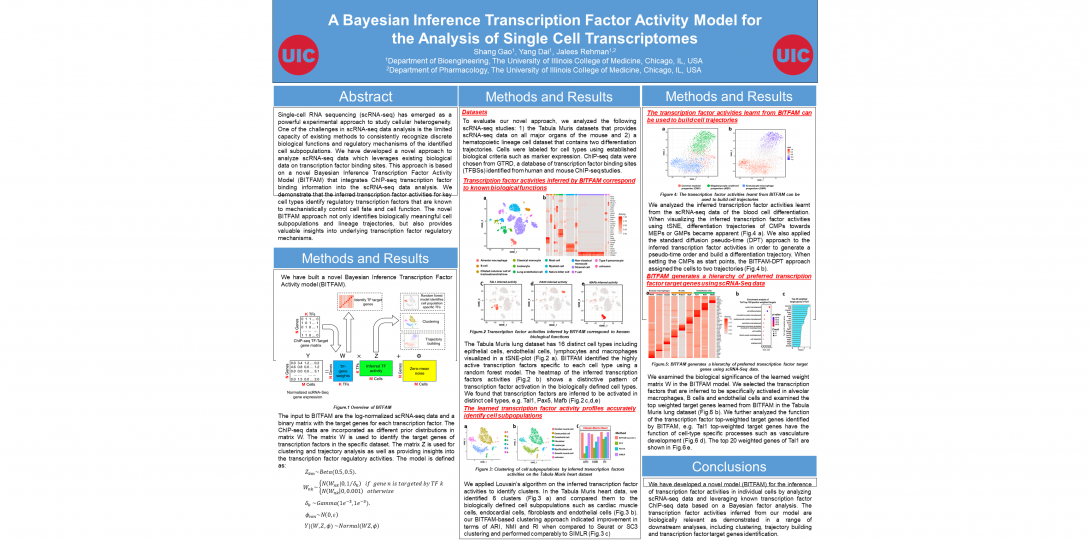
- BITFAM for inferring transcription factor activities from single cell RNA-seq data (Genome Research, 2021). Also read the UIC press about this tool.
- MeDEStrand an improved method to infer genome-wide absolute methylation levels from DNA enrichment data (BMC Bioinformatics, 2018)
B. Recent tools for microbiome-omics studies
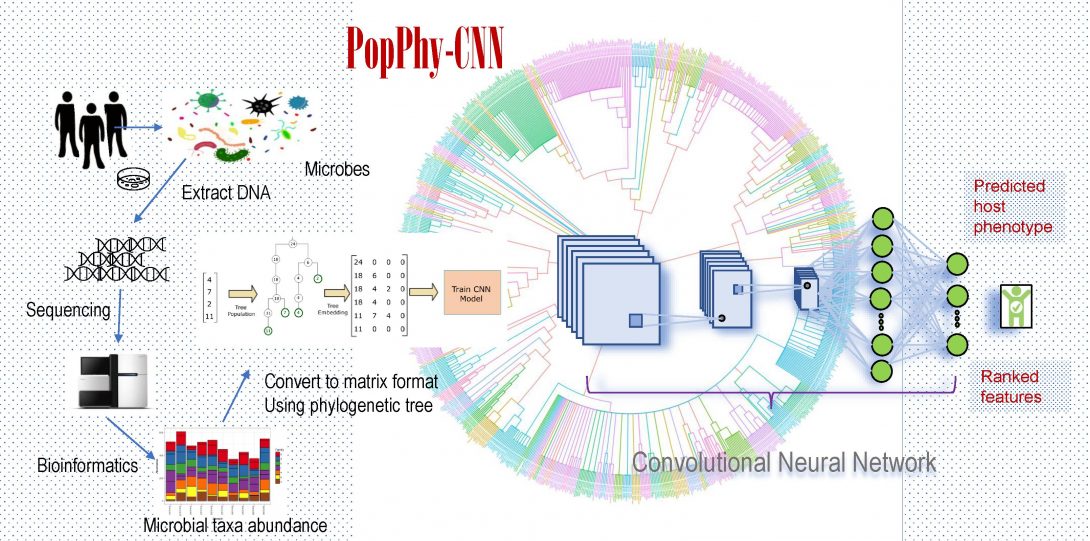
- PopPhy-CNN (IEEE-BHI, 2020) and Meta-Signer (F1000, 2021) for host phenotype prediction and microbial biomarker detection using metagenomics data
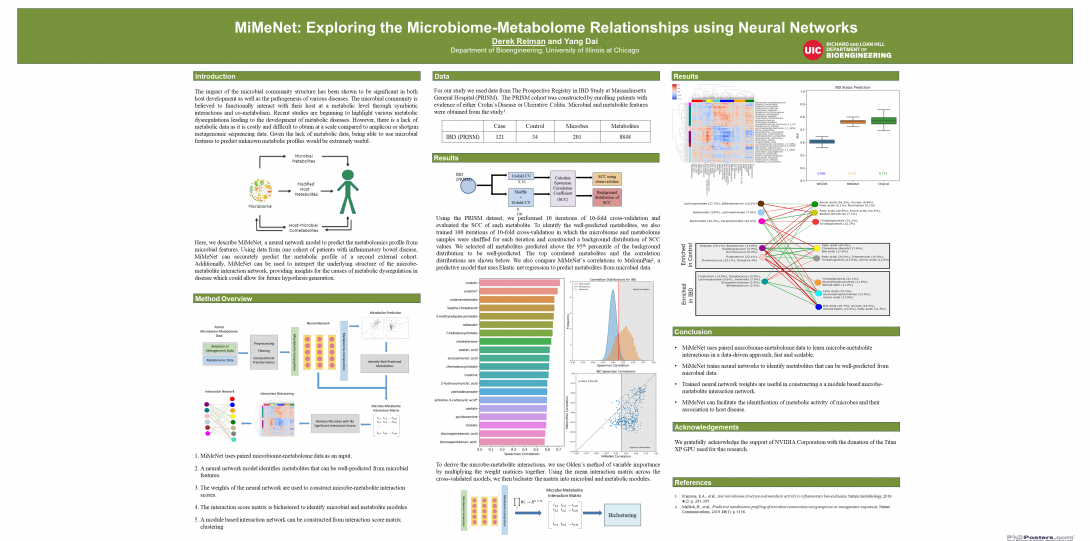
- MiMeNet for modeling microbiome-metabolome interaction by integrating microbiome and metabolome data (PLoS Computational Biology, 2021)
- MetaLonDA a R package for identifying time intervals of differentially abundant features in metagenomic longitudinal studies (Microbiome, 2018)
C. A database MOMMI-MP providing comprehensive analysis results of time-course metabolic/microbiome/metabolome profiles during mouse pregnancy (2023)
D. We collaborate with biomedical researchers on omics data analysis (funded by NIH/NSF)
- Drs. Bulun and Yin's Group at Dept. of Obstetrics & Gynecology, Northwestern University (JCI insight, Hum Reprod; Reproductive Biology; Stem Cell Reports; Endocrinology; PLoS Genet; PLoS One)
- Dr. Rehman's Group at Dept. Biomedical Engineering (Genome Research; JCI Insight; Elife; JCI Insight; BMC Bioinformatics)
- Dr. Khetani's Group at Dept. of Biomedical Engineering
- Dr. Koh's Group at Dept. of Kinesiology and Nutrition, College of Applied Public Health (J of Immunology)
- Dr. Peñalver Bernabé's group at Dept. Biomedical Engineering
- Dr. Layden's group at Division of Diabetes, Endocrinology and Metabolism, College of Medicine (Clin Transl Gastroenterol, Gastroenterology)
- Dr. Zhou and Dr. DiPietro at College of Dentistry (Scientific Reports; Int J Mol Sci; BMC Cancer; Mol Carcinog; J Biol Chem; Biochem J; Biomark Cancer)
- Finn and Perkins' Group at Dept. of Medicine (PLoS Computational Biology; Microbiome; PLoS One)